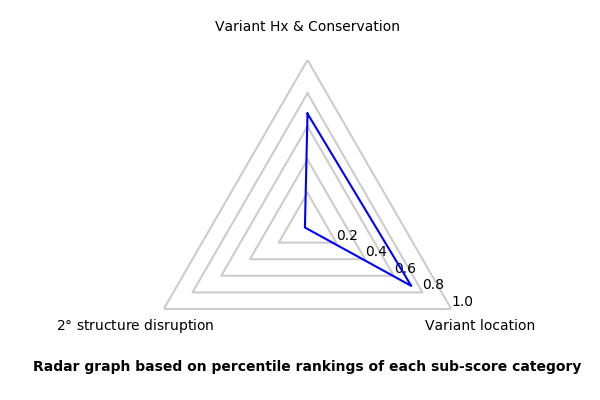
MitoTIP Sub-Scoring for Variant 15923G
MitoTIP Scoring Details
| rCRS Position | rCRS NT | Query NT | Numerical Scores | Percentile | Status | |||
|---|---|---|---|---|---|---|---|---|
| Variant Hx and Conservation | Variant Location | 2° Structure | Prediction | |||||
| 15923 | A | G | 10.279 | 2.167 | 0.000 | 12.446 | 46.60% | confirmed pathogenic * |
GenBank Frequency Information
| L lineages African |
M lineages Asian |
N lineages Eurasian |
Highest hg lineage(s) |
|---|---|---|---|
| 0.00% ( 0 / 6836 ) | 0.00% ( 0 / 12945 ) | 0.00% ( 0 / 42735 ) | NA |